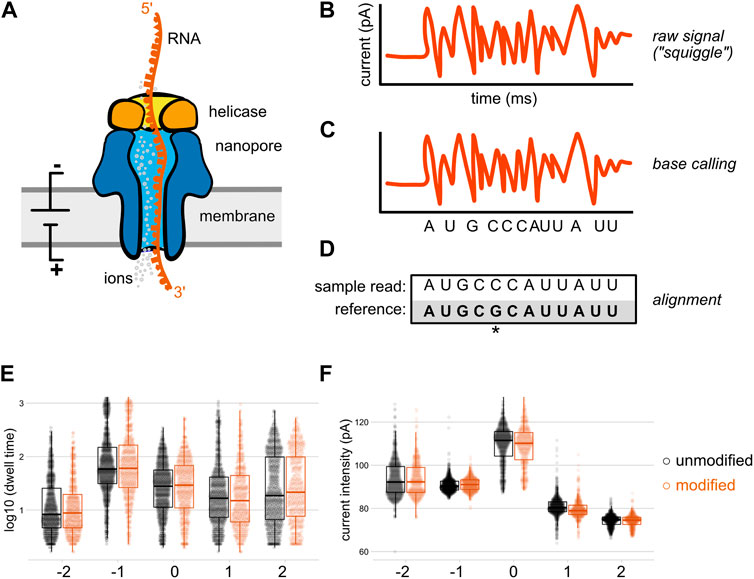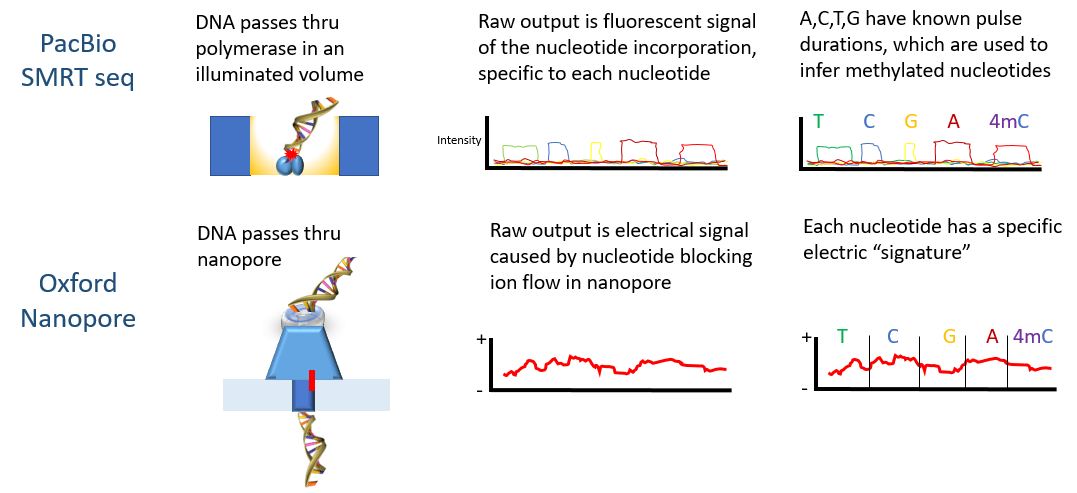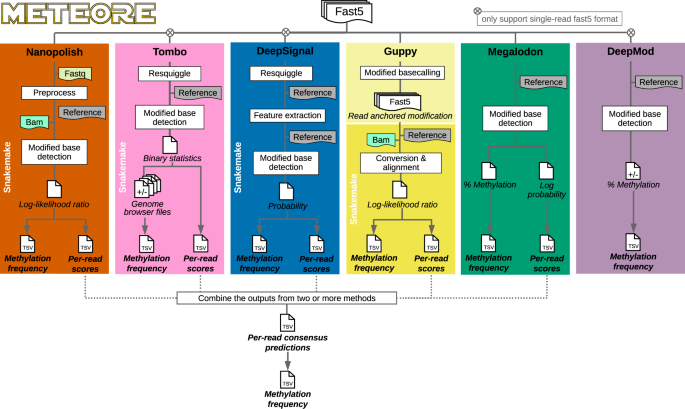
Systematic benchmarking of tools for CpG methylation detection from nanopore sequencing | Nature Communications

Methylation calling from nanopore data and comparison to gold standard... | Download Scientific Diagram
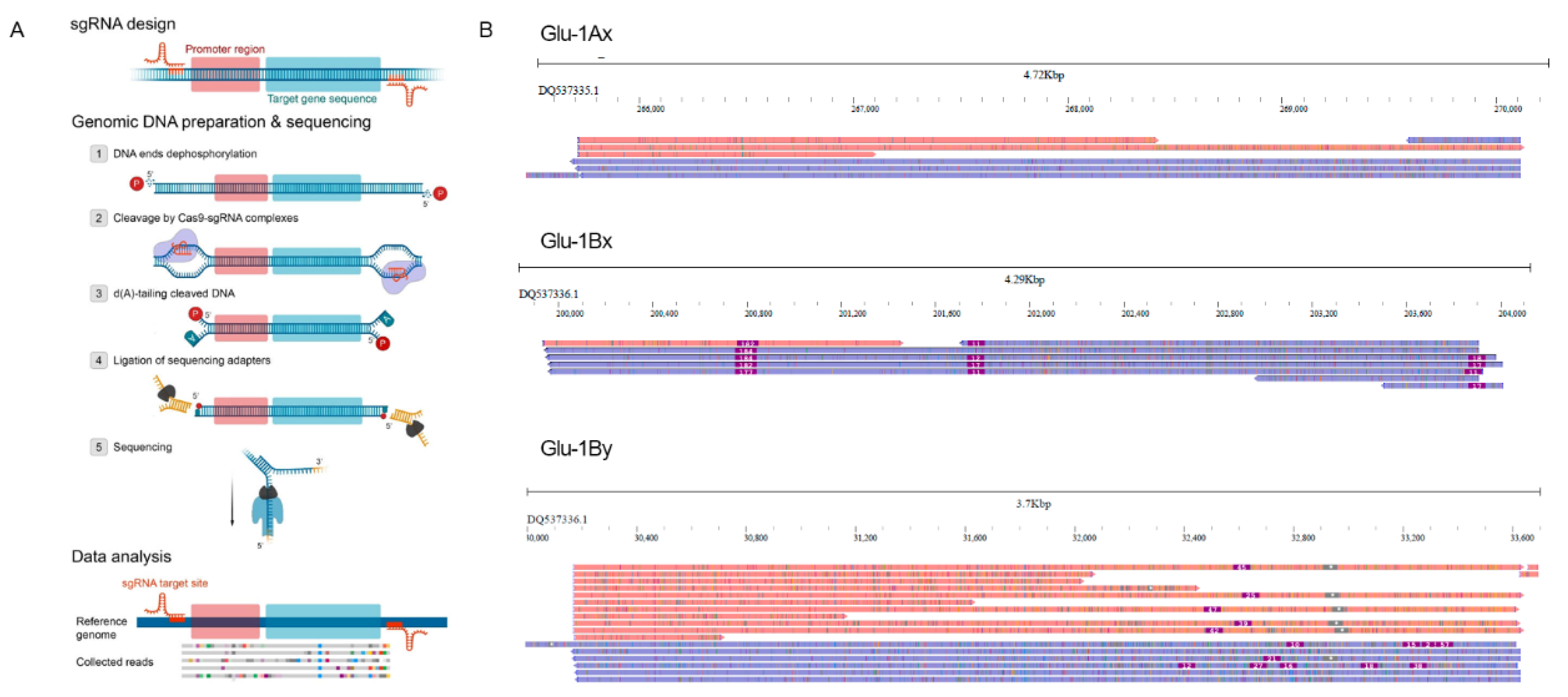
Plants | Free Full-Text | Searching for a Needle in a Haystack: Cas9-Targeted Nanopore Sequencing and DNA Methylation Profiling of Full-Length Glutenin Genes in a Big Cereal Genome
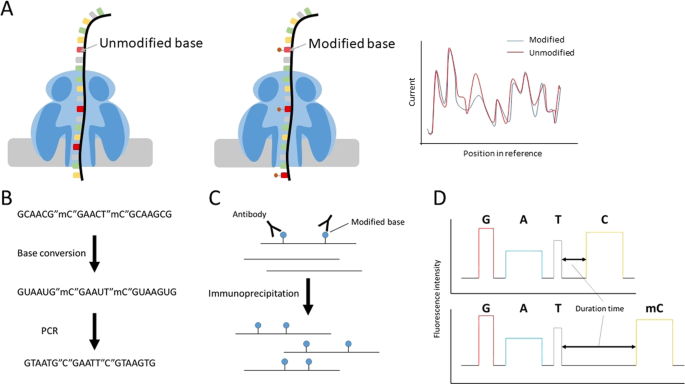
Recent advances in the detection of base modifications using the Nanopore sequencer | Journal of Human Genetics

Discovering and exploiting multiple types of DNA methylation from individual bacteria and microbiome using nanopore sequencing | bioRxiv
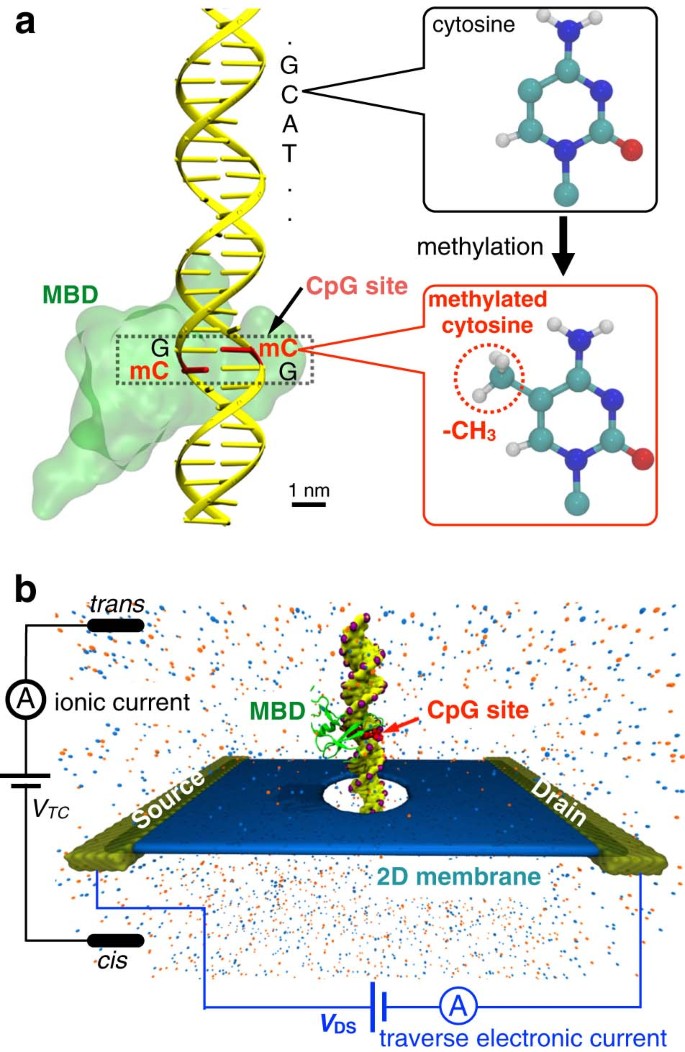
Detection and mapping of DNA methylation with 2D material nanopores | npj 2D Materials and Applications

Beyond sequencing: machine learning algorithms extract biology hidden in Nanopore signal data: Trends in Genetics
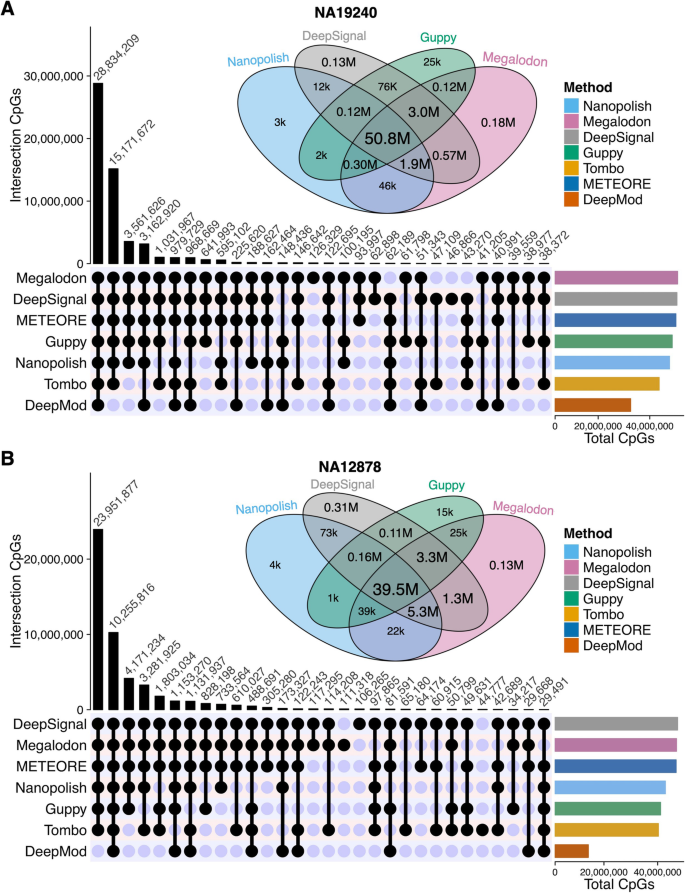
DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation | Genome Biology | Full Text
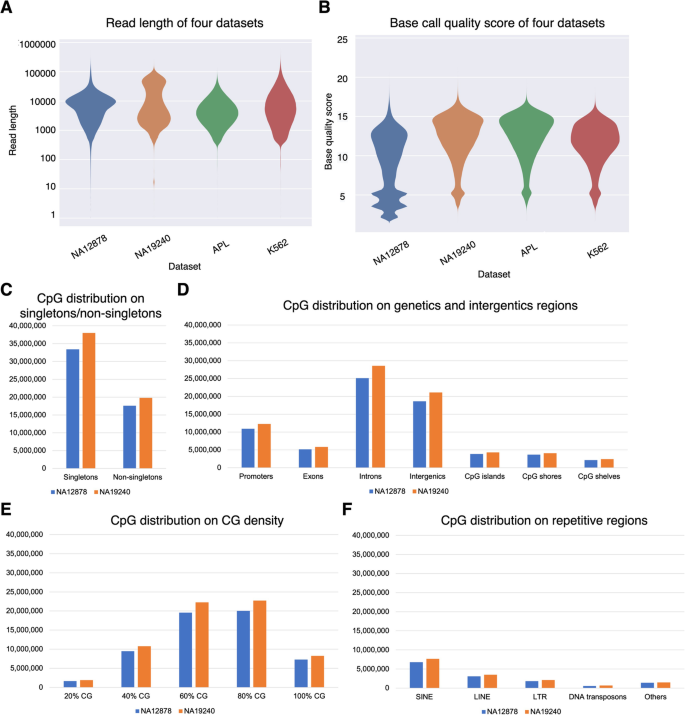
DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation | Genome Biology | Full Text
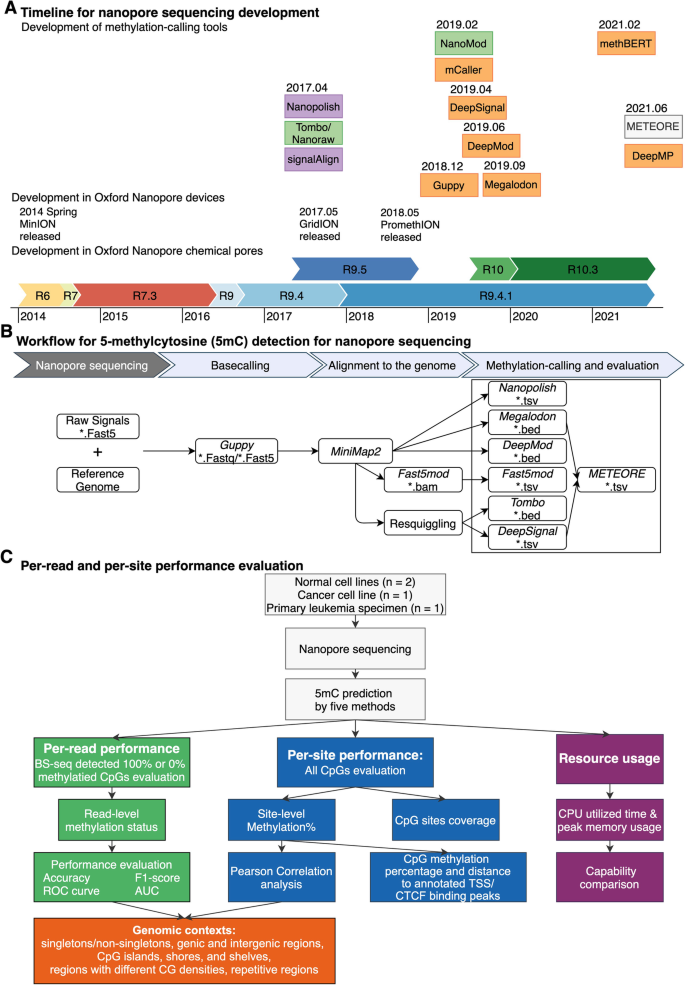
DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation | Genome Biology | Full Text
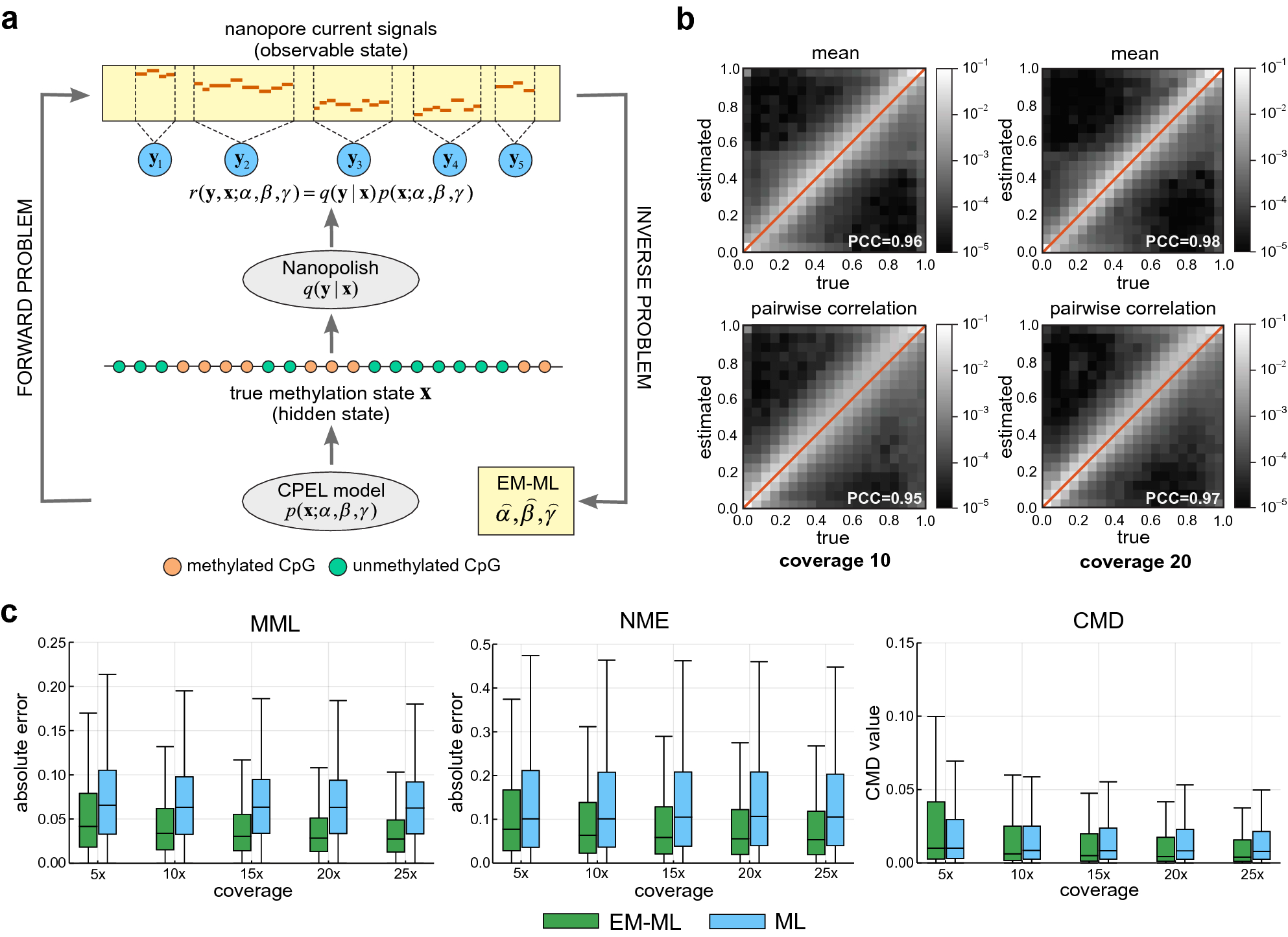
Estimating DNA methylation potential energy landscapes from nanopore sequencing data | Scientific Reports
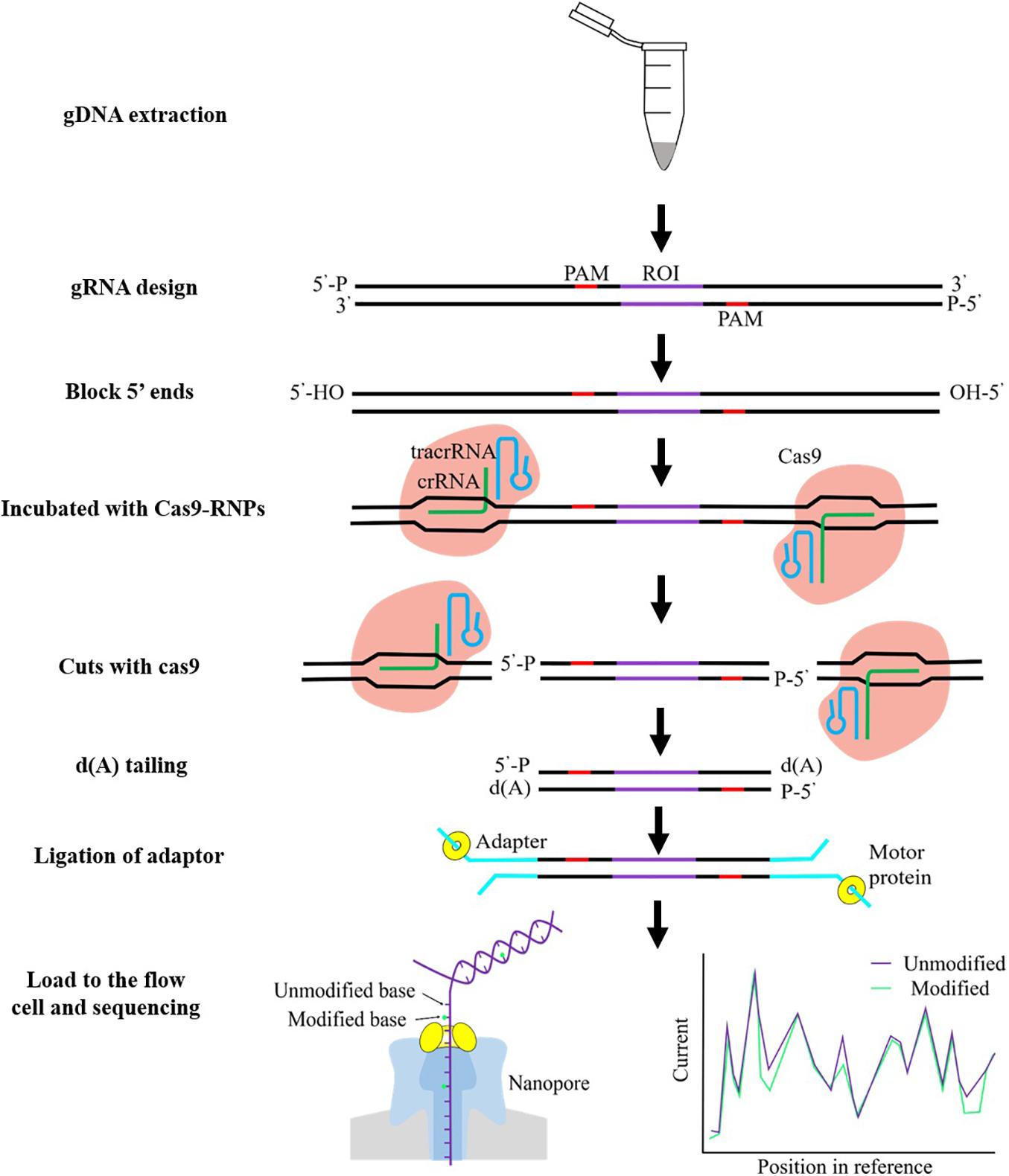
Frontiers | Cancer Biomarkers Discovery of Methylation Modification With Direct High-Throughput Nanopore Sequencing